Discover the world of
BD Multi-Omics
The Multi-Omics Alliance
Born to transform
We are pleased to announce the formation of the “BD Multi-Omics Alliance”, consisting of a balanced team of 7 renowned academic experts from across Europe and a group of committed BD employees. The Alliance has taken on the challenge to evaluate the scientific impact of the BD Multi-Omics Technologies in basic and translational research through its use in real-life applications defined by the experts and relevant to a broad scientific community.
Want to learn more about how Multi-Omics is helping science?
Watch our teaser videos to get a taste of how scientists are using this exciting tool to answer new questions!
Watch our BD single-cell Multi-Omics teaser videos series now and register to get the full webinars !
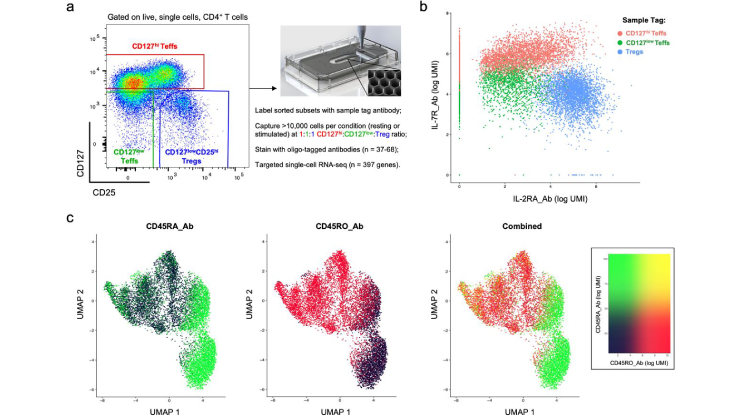
Simultaneous single-cell mRNA and epitope mapping of human CD4+ T-cells using the BD Rhapsody™ and the BD™ AbSeq platform
Simultaneous single-cell mRNA and epitope mapping of human CD4+ T-cells using the BD Rhapsody™ and the BD™ AbSeq platform.
Dr Ricardo C. Ferreira, University of Oxford
Evolution, challenges and the future of single-cell immunology
Description of the single-cell Multi Omics workflow that includes sample preparation, single-cell isolation and molecular barcoding, library preparation, sequencing and data analysis.
Christina Chang, PhD
Testimonial
Prof. Dr. Holger Garn/Kim Pauck Institute of Laboratory Medicine and Pathobiochemistry Philipps University of Marburg, Germany
“We were impressed with the straightforward workflow of the whole procedure of BD Rhapsody™ system from sample preparation to data analysis. Furthermore, when handling very small tissues (we used single mouse lymph nodes in our case), the BD Rhapsody™ is able to generate a multitude of in-depth information without waste of precious ex vivo material. Even without any prior experience in single-cell sequencing, we didn’t stumble on a single step with our first run and generated a massive data set, which was even workable for someone with only basic bioinformatics knowledge. The pipeline BD provides for processing the sequencing raw data is clearly arranged and easy to pick up. Data analysis with SeqGeq provides an excellent starting point for people already familiar with FlowJo and is a joy to work with. The ability to combine a massive amount of surface markers with targeted gene analysis on a single cell level while retaining the option to multiplex for a clean comparison between samples of different origin is a very strong method, which we certainly did not regret to pick up for our lab.”
Measuring up to new single cell challenges with the Rhapsody™

Alison Devonshire, Science Leader (Nucleic Acid Metrology)
As Alison Devonshire, Science Leader, Nucleic Acid Metrology, noted, "This new technology will allow us to further develop our single cell analysis capabilities to support our activities within engineering biology and precision medicine, for example for stem cell-based therapies or understanding the heterogeneity of cancer cells.”
Taken from on www.lgcgroup.com
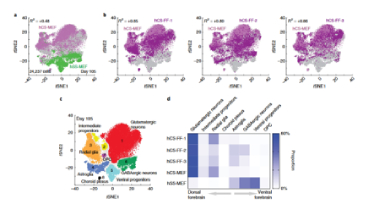
Discover our latest publications on Multi-Omics :
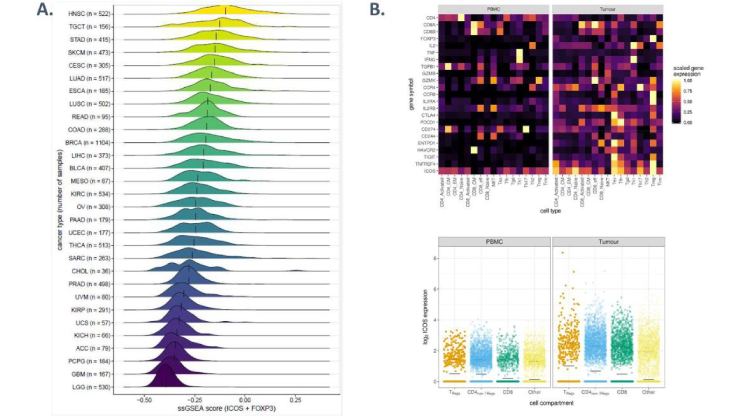
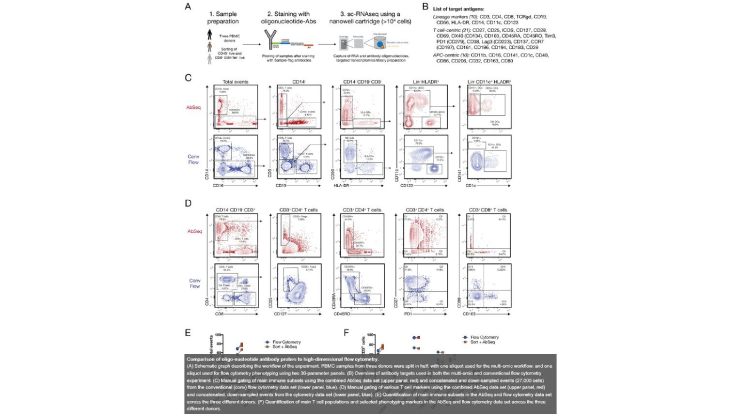
A targeted multi-omic analysis approach measures protein expression and low abundance transcripts on the single cell level
Florian Mair, Jami R. Erickson, Valentin Voillet, Yannick Simoni, Timothy Bi, Aaron J. Tyznik, Jody Martin, Raphael Gottardo, Evan W. Newell, Martin Prlic
High throughput single-cell RNA sequencing (sc-RNAseq) has become a frequently used tool to assess immune cell function and heterogeneity. Recently, the combined measurement of RNA and protein expression by sequencing was developed, which is commonly known as CITE-Seq. Acquisition of protein expression data along with transcriptome data resolves some of the limitations inherent to only assessing transcript, but also nearly doubles the sequencing read depth required per single cell. Furthermore, there is still a paucity of analysis tools to visualize combined transcript-protein datasets.
BD Rhapsody™ platform
Analyze the expression of hundreds of genes across tens of thousands of single cells in parallel
BD™ AbSeq oligos
Deepen your understanding and accelerate your research with the BD™AbSeq multiomic assay for single-cell analysis
New Release : WTA
Whole Transcriptome Analysis Amplification Kit is now available !
Interested in Multi-Omics ? Let’s keep in touch